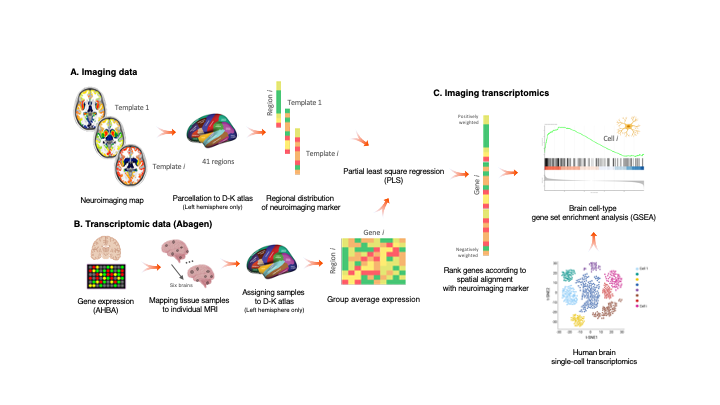
Imaging Transcriptomics
Imaging transcriptomics is a methodology that allows to identify patterns of correlation between gene expression and some property of brain structure or function as measured by neuroimaging (e.g., MRI, fMRI, PET).
The imaging-transcriptomics package allows performing imaging transcriptomics analysis on a neuroimaging scan (e.g., PET, MRI, fMRI...).
The software is implemented in Python3 (v.3.7), its source code is available on GitHub, it can be installed via Pypi and is released under the GPL v3 license.
NOTE Versions from v1.0.0 are or will be maintained. The original script linked by the BioRxiv preprint (v0.0) is still available on GitHub but no changes will be made to that code. If you have downloaded or used that script please update to the newer version by installing this new version.
Installation
NOTE We recommend to install the package in a dedicated environment of your choice (e.g., venv or anaconda). Once you have created your environment and you have activated it, you can follow the below guide to install the package and dependencies. This process will avoid clashes between conflicting packages that could happen during/after the installation.
To install the imaging-transcriptomics Python package, first you will need to install a packages that can't be installed directly from PyPi, but require to be downloaded from GitHub. The package to install is pypyls. To install this package you can follow the installation on the documentation for the package or simply run the command
pip install -e git+https://github.com/netneurolab/pypyls.git/#egg=pyls
to download the package, and its dependencies directly from GitHub by using pip.
Once this package is installed you can install the imaging-transcriptomics package by running
pip install imaging-transcriptomics
Usage
Once installed the software can be used in two ways:
- as standalone script
- as part of some python script
WARNING Before running the script make sure the Pyhton environment where you have installed the package is activated.
Standalone script
To run the standalone script from the terminal use the command:
imagingtranscriptomics options
The options available are:
-i (--input): Path to the imaging file to analise. The path should be given to the program as an absolute path (e.g.,/Users/myusername/Documents/my_scan.nii, since a relative path could raise permission errors and crashes. The script only accepts imaging files in the NIfTI format (.nii,.nii.gz).-v (--variance): Amount of variance that the PLS components must explain. This MUST be in the range 0-100.NOTE: if the variance given as input is in the range 0-1 the script will treat this as 30% the same way as if the number was in the range 10-100 (e.g., the script treats the inputs
-v 30and-v 0.3in the exact same way and the resulting components will explain 30% of the variance).-n (--ncomp): Number of components to be used in the PLS regression. The number MUST be in the range 1-15.--corr: Run the analysis using Spearman correlation instead of PLS.NOTE: if you run with the
--corrcommand no other input is required, apart from the input scan (-i).-o (--output)(optional): Path where to save the results. If none is provided the results will be saved in the same directory as the input scan.
WARNING: The
-iflag is MANDATORY to run the script, and so is one, and only one, of the-nor-vflags. These last two are mutually exclusive, meaning that ONLY one of the two has to be given as input.
Part of Python script
When used as part of a Python script the library can be imported as:
import imaging_transcriptomics as imt
The core class of the package is the ImagingTranscriptomics class which gives access to the methods used in the standalone script. To use the analysis in your scripts you can initialise the class and then simply call the ImagingTranscriptomics().run() method.
import numpy as np
import imaging_transcriptomics as imt
my_data = np.ones(41) # MUST be of size 41
# (corresponds to the regions in left hemisphere of the DK atlas)
analysis = imt.ImagingTranscriptomics(my_data, n_components=1)
analysis.run()
# If instead of running PLS you want to analysze the data with correlation you can run the analysis with:
analysis.run(method="corr")
Once completed the results will be part of the analysis object and can be accessed with analysis.gene_results.
The import of the imaging_transcriptomics package will import other helpful functions for input and reporting. For a complete explanation of this please refer to the official documentation of the package.
Documentation
The documentation of the script is available at imaging-transcriptomics.rtfd.io/.
Troubleshooting
For any problems with the software you can open an issue in GitHub or contact the maintainer) of the package.
Citing
If you publish work using imaging-transcriptomics as part of your analysis please cite:
Imaging transcriptomics: Convergent cellular, transcriptomic, and molecular neuroimaging signatures in the healthy adult human brain. Daniel Martins, Alessio Giacomel, Steven CR Williams, Federico Turkheimer, Ottavia Dipasquale, Mattia Veronese, PET templates working group. bioRxiv 2021.06.18.448872; doi: https://doi.org/10.1101/2021.06.18.448872
Imaging-transcriptomics: Second release update (v1.0.2).Alessio Giacomel, & Daniel Martins. (2021). Zenodo. https://doi.org/10.5281/zenodo.5726839